Volume 6 (2023) Issue 3 No.4 Pages 80-86
Abstract
The most important prognostic factor for Philadelphia chromosome-positive acute lymphoblastic leukemia (
Introduction
Treatment outcomes for Philadelphia chromosome-positive acute lymphoblastic leukemia (
Minimal residual disease (MRD) is the most powerful and important prognostic factor of ALL6. However, it has been evaluated during treatment. Negative measurable MRD at the end of the initial induction chemotherapy is a reliable and good prognostic factor7–9. However, there are cases where ALL relapses, although MRD disappears early, and cases where ALL does not relapse, even if MRD persists before allo-HSCT. If genetic mutations in leukemia cells at the initial stage of the disease are identified as risk factors, in addition to MRD, the selection of optimal treatment may be possible. In addition, allo-HSCT during the first phase of
Furthermore, if Ph+ALL with poor allo-HSCT results could be extracted at the time of disease onset, it would be helpful to optimize chemotherapy until allo-HSCT is performed or transplantation methods are considered. In this regard, risk assessment at disease onset is considered more important than MRD because MRD is assessed during treatment. In this study, we believe gene mutation analysis at the beginning of the disease might be an effective prognostic factor other than MRD; hence, we conducted a retrospective analysis at a single center.
Materials and Methods
Patients
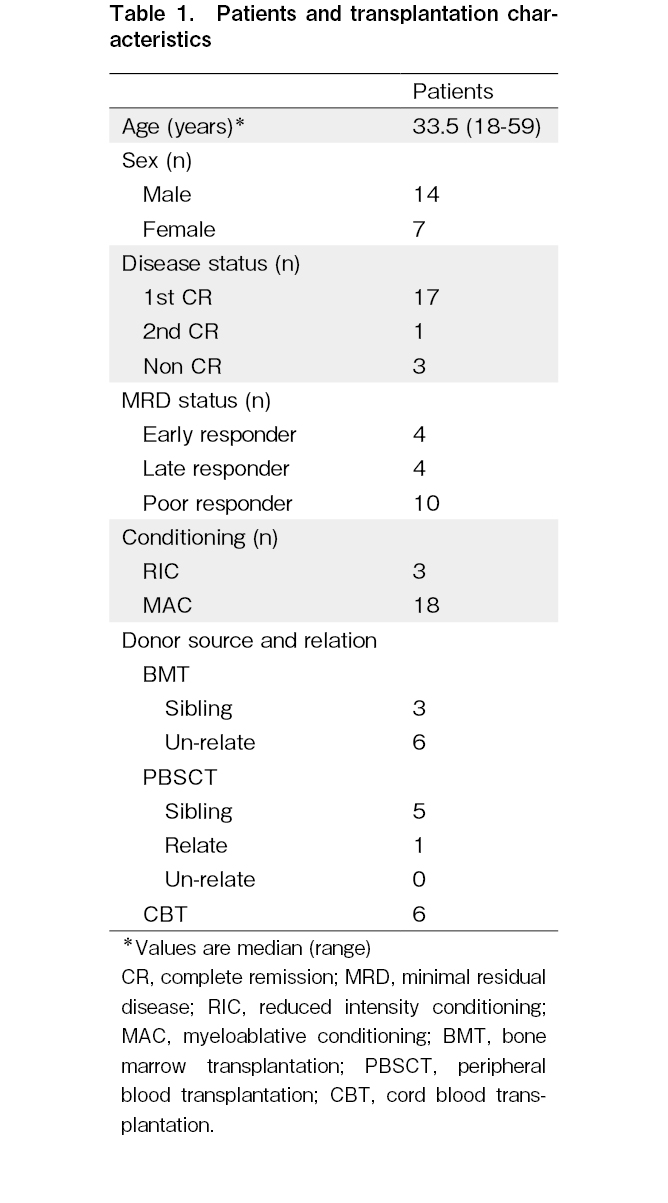
Among the Ph+ALL cases in which allo-HSCT was performed at our institution between December 2006 and October 2021, 21 cases where leukemia cells were preserved at the time of onset or recurrence were analyzed in this study. The observation period was from March 2022. Chemotherapy before transplantation included hyper-CVAD (fractionated cyclophosphamide, vincristine, adriamycin, and dexamethasone) therapy in three patients and Japan Adult Leukemia Study Group (JALSG) multidrug chemotherapy in 18 patients.
Of the 21 patients, 11 received imatinib, and 10 received dasatinib. Pre-transplant treatment included three cases of reduced-intensity conditioning (RIC) and 18 cases of myeloablative conditioning (MAC). For RIC, we used fludarabine+melphalan±total body irradiation (TBI) of 4 Gy in 3 cases. For MAC, we used cyclophosphamide (CY)+TBI of 12 Gy in 10 cases, etoposide (VP)+CY+TBI of 12 Gy in 7 cases, and cytarabine (CA)+CY+TBI of 10 Gy in 1 case. For graft-versus-host disease prophylaxis, cyclosporine (CsA) and short-term methotrexate (sMTX) were administered to HLA-matched siblings, and tacrolimus (Tac)+sMTX was used as an unrelated donor source. In one case, with the mother as the donor, Tac+sMTX was used because the HLA disparity was 4/6. The study was conducted in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) and approved by the Institutional Review Board of Tokai University Hospital (10I-61/12I-09).
Sample collection and methods
Mononuclear cells were isolated from the nucleated bone marrow cells at the time of initial onset and stored in liquid nitrogen until analysis. DNA was extracted from stored samples (Qiagen DNA Blood Kit; Qiagen Inc., Redwood City, CA, USA) and adjusted to a Tris-EDTA buffer concentration of 50 ng/L. According to the MLPA kit protocol (MRC Holland, Amsterdam, Netherlands), denaturation, hybridization, ligation, and polymerase chain reaction (PCR) were performed in a thermal cycler and analyzed using a capillary sequencer. As in the previous report, a probe ratio between 0.75 and 1.3 was within the normal range. A probe ratio
MRD measurements were evaluated as the first MRD immediately before consolidation therapy after remission induction therapy and were performed 2-4 weeks before allo-HSCT. The first MRD measurement was quantitative using real-time PCR or qualitative using nested PCR. The second measurement before transplantation was performed using nested PCR9. The second PCR product was confirmed using electrophoresis and defined as MRD-negative when no minor or major BCR-ABL bands were identified. Regarding MRD status, the first and second negatives were designated as early responders, the first positive and second negative were designated as late responders, and the second positive or non-remission was designated as poor responders.
Statistics analyses
All categorical variables were compared using Fisher's exact test, including patient characteristics, disease status, transplantation characteristics, and MRD status. Quantitative variables, such as age, white blood cell (WBC) count, and blast ratio, were compared using t-tests. Time-to-event analyses were performed using the Cox regression model to determine the association between gene deletion and overall survival (OS) and disease-free survival (DFS). Other clinical features such as age, WBC count, blast count at diagnosis, and MRD status were also analyzed using the Cox regression model to determine the relationship between OS and DFS.
Gene deletions, other clinical features, and relapse were analyzed using a competing risk regression model to determine death without recurrence as a competing event. MLPA and post-transplantation results were evaluated for IKZF1, CDKN2A/2B, PAX5, and IKZF1+ abnormalities. The number of gene deletions (0-6) was classified and evaluated as 0, 1, 2, 3, and more. Relapse was considered a hematological recurrence, whereas molecular genetic recurrence was not a recurrence event. Statistical significance was defined as p<0.05. All statistical analyses were performed using Stata software version 16 (College Station, TX, USA).
Results
Outcomes of transplantation
The transplantation information for each patient is presented in Table 1. The median follow-up period for all patients was 1,022 days (2.8 years), ranging from 30 months to 14.7 years. On the last day of the observation period, 11 of the 21 patients survived, 2 had primary disease deaths, and 8 had transplant-related deaths. Pre-transplant disease status was observed in 18 patients in remission and 3 in non-remission. Additionally, 4 patients were negative in the initial MRD measurement, and 17 were positive. The second MRD measurement before allotransplantation showed 8 negative and 10 positive cases. Therefore, the MRD status was as follows: 4 cases of early responders, 4 of late responders, 3 not in complete remission (non-CR), and 10 poor responders. OS, DFS, and recurrence rates were assessed according to age, initial WBC count, initial blast count, and MRD status, none of which had a significant effect (age: P=0.61, P=0.97, and P=0.87; WBC count: P=0.68, P=0.96, and P=0.67; blast count: P=0.70, P=0.99, and P=0.069; MRD: P=0.82, P=0.96, and P=0.75). Furthermore, 3 patients had significantly worse OS and DFS [hazard ratio (HR), 11.3; 95% confidence interval (CI), 1.8-72.5, P=0.010 and HR, 14.9; 95% CI, 2.1-107.3, P=0.007]; all these patients died from transplant-related complication without relapse of disease.
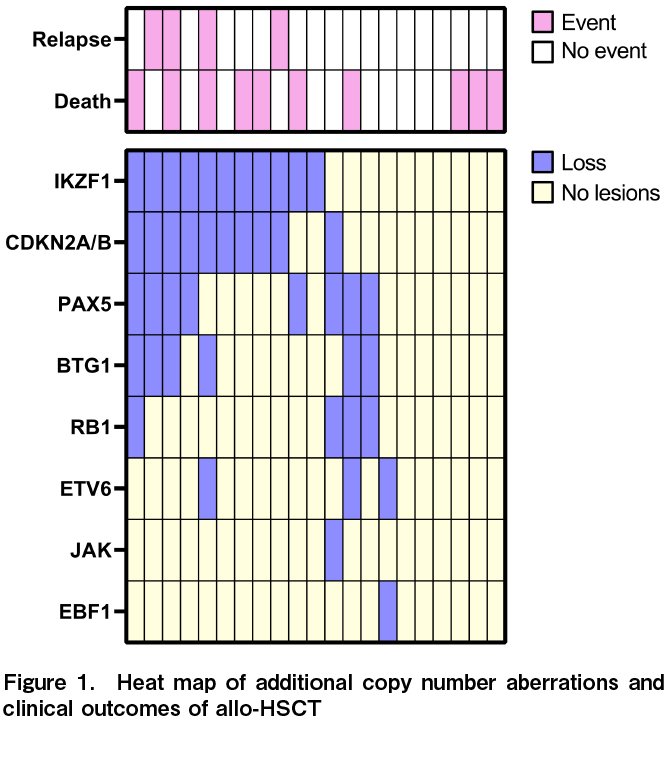
MLPA analysis
MLPA tests showed that 71.4% of patients (15/21) had at least one abnormality involving IKZF1,
Transplantation outcomes and MLPA results
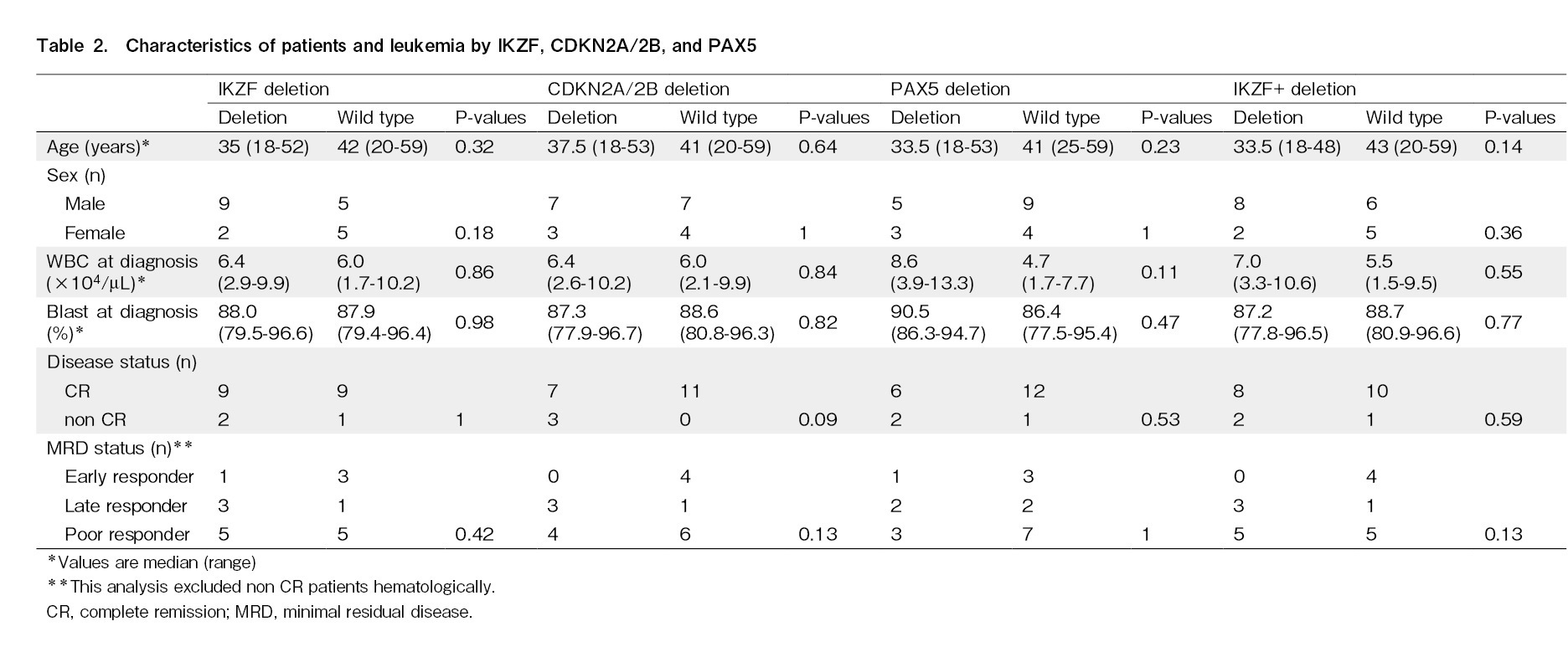
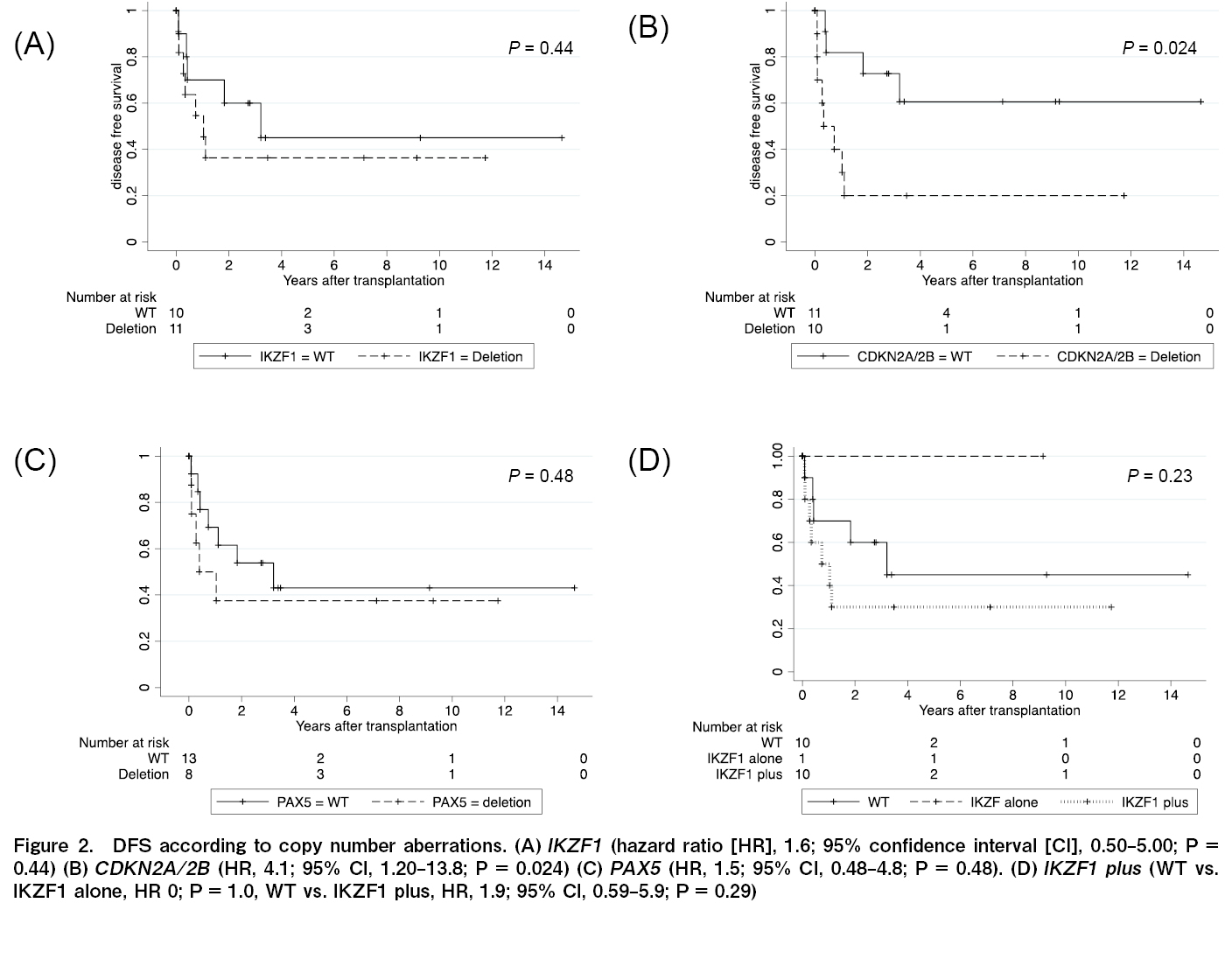
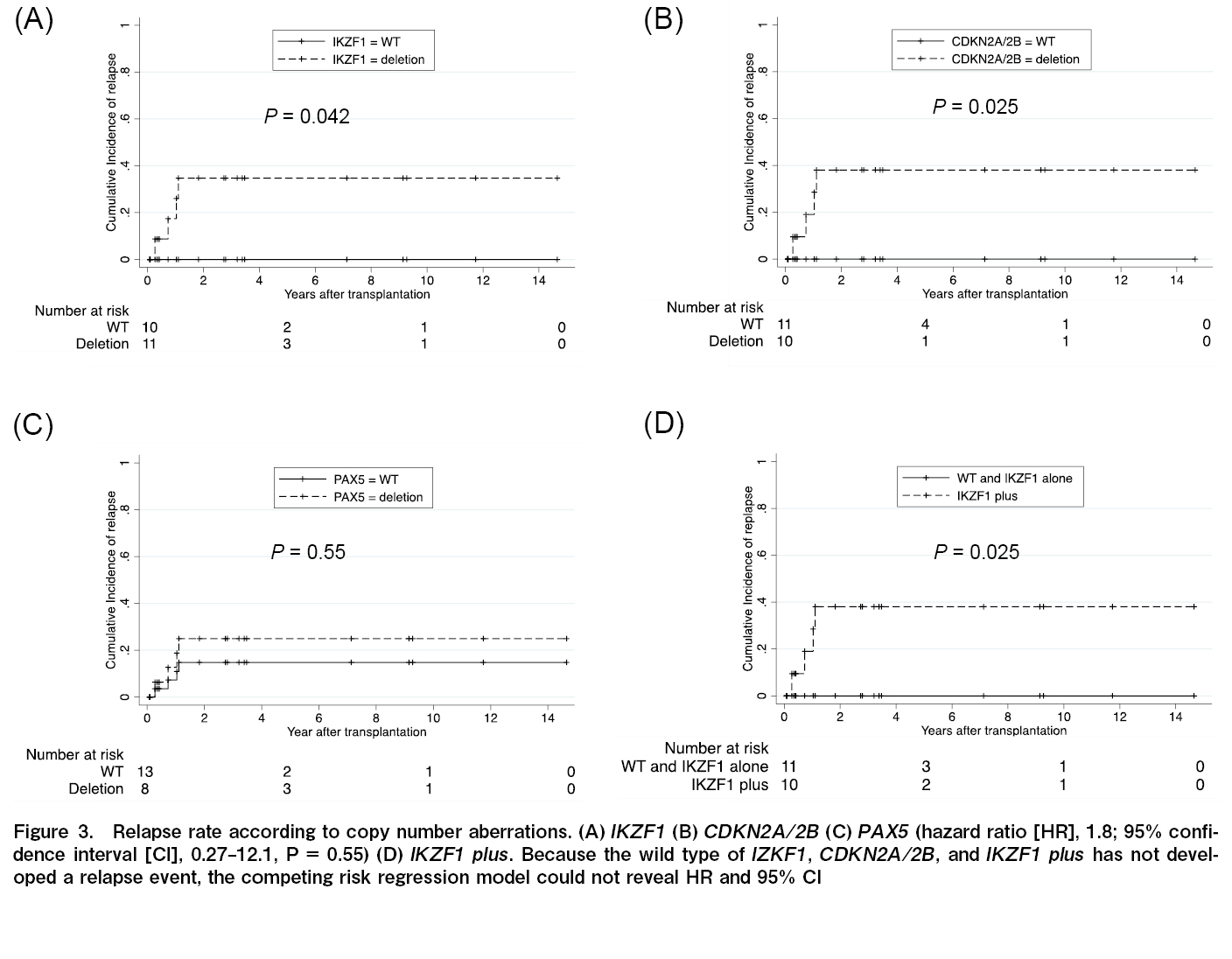
The effects of IKZF1, CDKN2A/2B, and PAX5 deletions on the OS, DFS, and recurrence rates were analyzed. Similar analyses were performed for IKZF1+ and all seven gene copy number variations. Gene defects that significantly affected OS were not observed in any of the analyses (IKZF1; P=0.75; CDKN2A/2B; P=0.25; PAX5; P=0.82; JAK; P=0.056; ETV6; P=0.12; RB1; P=0.086; BTG1; P=0.32; and EBF1; P=1.0). The DFS rate was significantly lower in patients with CDKN2A/2B deletion only than wild-type CDKN2A/2B (HR, 4.1; 95% CI, 1.20-13.8, P=0.024) (Figure 2). Relapse rates were significantly higher for than each wild-types
Next, the relative CDKN2A/2B deletion, DFS, and relapse rates were analyzed (excluding the four non-CR cases). In these 18 cases, CDKN2A/2B deletion was not significantly associated with DFS (P=0.098). However, the deletion was significantly associated with the relapse rate (P=0.0032).
Despite the late or poor MRD response, no patient had relapsed leukemia when genetic aberrations in IKZF1 or CDKN2A/2B were wild-type. The MRD status at allo-HSCT was not significantly associated with relapse after transplantation (P=0.75).
Discussion
Although the number of cases was small, the treatment outcomes of allo-HSCT for Ph+ALL at a single institution were analyzed based on gene defect analysis using MLPA. DFS and recurrence rates were significantly higher in patients with CDKN2A/2B deficiency, and recurrence rates were significantly higher in cases with IKZF1 and IKZF1+ gene defects. In this cohort, the presence or absence of MRD immediately before transplantation did not affect the transplant outcomes, suggesting that CDKN2A/2B deficiency may be a stronger prognostic factor than MRD deficiency.
The results of allo-HSCT for Ph+ALL have shown that the presence or absence of CDKN2A/2B deficiency affects DFS more strongly than IKZF1 deficiency3. Furthermore, IKZF1 deficiency did not affect prognosis in adults with Ph+ALL and Ph-ALL disease11. Moreover, IKZF1+ with CDKN2A/2B or PAX5 deficiency is a poor prognostic factor compared to IKZF1 alone, suggesting that CDKN2A/2B deficiency is an important prognostic factor5. Although this study was conducted at a single institution, the same trend as that previously reported was confirmed for allo-HSCT for Ph+ALL in Japan.
CDKN2A encodes p16 (INK4a) and p14 (ARF), and INK4a controls cell cycle progression by blocking the activities of CDK4 and CDK6. ARF is a physiological inhibitor of MDM2, an E3 ubiquitin ligase that controls the activity and stability of P5312. CDKN2B encodes a cyclin-dependent kinase inhibitor that forms a complex with CDK4 or CDK6 and prevents the activity of CDK, controlling cell cycle progression through the G1 phase12. In ALL, CDKN2A/2B deletion is associated with a high WBC count at the first onset, advanced age at diagnosis, and Ph-like ALL13, 14. Thus, CDKN2A/2B deletion may be an important prognostic factor of ALL.
This study aimed to determine whether genetic defect analysis at the onset of Ph+ALL could be used to determine transplantation indications. Most post-transplant recurrences of Ph+ALL occur within 2 years of transplantation1. In this regard, the observation period in our study was 2.5-14.7 years, and we believe that we have obtained a sufficient analysis period to confirm the relapse. Moreover, there was no single recurrence in this analysis but rather transplant-related death that led to reduced survival rates among approximately 30% of the cases in whom the MLPA method did not detect a gene defect. These patients may be candidates for cases where allotransplantation can be avoided. However, we could not observe recurrence due to NRM. Thus, our results should be interpreted with caution owing to the small number of cases analyzed.
The limitation of this analysis was the small number of cases; therefore, we did not evaluate the relation OS, DFS and MRD, which has the most significant impact on treatment outcomes after allo-HSCT of ALL. Previous study findings, and our results, have shown that early MRD negativity is a positive prognostic factor9, 15. The importance of MRD was not demonstrated in our study cohort, which included only cases in which samples for MLPA analysis were obtained, owing to the small number of cases.
In contrast, there were no cases of recurrence in MRD-positive poor responders before allo-HSCT in patients without IKZF1 or CDKN2A/2B defects. Owing to the small cohort size, other gene defects might not significantly affect the transplant outcome. Conversely, the fact that CDKN2A/2B is a factor affecting prognosis, despite the small number of cases, indicates that this gene defect is a poor prognostic factor, as shown in prior reports3, 5. Because Ph+ALL treatment has changed markedly owing to the advent of molecular-targeted drugs, we believe that a meaningful analysis is possible in this cohort analysis, which has a relatively uniform treatment history.
In this study, we confirmed that gene defect analysis using MLPA effectively predicted the prognosis at the onset of Ph+ALL in a single-center Japanese cohort. As the number of cases increases in the future, gene defect analysis using the MLPA method can help effectively select cases in which allogeneic transplantation can be avoided, together with MRD information.
Author Contributions
MO, EK, and KA conceived and designed the study; MO, SM, MT, RS, DO, YA, JA, KH, RH and SS collected samples and clinical data; MO and EK performed laboratory assessments; MO and KA performed statistical analyses; MO, YO, HK, and KA interpreted the data; MO, EK, and KH wrote the manuscript and created the figures and tables.
Conflicts of Interest
The authors declare no conflict of interest. Disclosure forms provided by the authors are available on the website.
References
1.Mizuta S, Matsuo K, Nishiwaki S, Imai K, Kanamori H, Ohashi K, et al. Pretransplant administration of imatinib for allo-HSCT in patients with BCR-ABL-positive acute lymphoblastic leukemia. Blood. 2014; 123: 2325-32.
2.Nishiwaki S, Akahoshi Y, Mizuta S, Shinohara A, Hirabayashi S, Noguchi Y, et al. Measurable residual disease affects allogeneic hematopoietic cell transplantation in Ph+ ALL during both CR1 and CR2. Blood Adv. 2021; 5: 584-92.
3.Pfeifer H, Raum K, Markovic S, Nowak V, Fey S, Obländer J, et al. Genomic CDKN2A/2B deletions in adult Ph+ ALL are adverse despite allogeneic stem cell transplantation. Blood. 2018; 131: 1464-75.
4.Gu Z, Churchman ML, Roberts KG, Moore I, Zhou X, Nakitandwe J, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet. 2019; 51: 296-307.
5.Kantarjian H, Stein A, Gökbuget N, Fielding AK, Schuh AC, Ribera JM, et al. Blinatumomab versus chemotherapy for advanced acute lymphoblastic leukemia. N Engl J Med. 2017; 376: 836-47.
6.Berry DA, Zhou S, Higley H, Mukundan L, Fu S, Reaman GH, et al. Association of minimal residual disease with clinical outcome in pediatric and adult acute lymphoblastic leukemia: A Meta-analysis. JAMA Oncol. 2017; 3: e170580.
7.Conter V, Bartram CR, Valsecchi MG, Schrauder A, Panzer-Grümayer R, Möricke A, et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: Results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood. 2010; 115: 3206-14.
8.Wood B, Wu D, Crossley B, Dai Y, Williamson D, Gawad C, et al. Measurable residual disease detection by high-throughput sequencing improves risk stratification for pediatric B-ALL. Blood. 2018; 131: 1350-9.
9.Hara R, Onizuka M, Kikkawa E, Shiraiwa S, Harada K, Aoyama Y, et al. Association between measurable residual disease kinetics and outcomes of Philadelphia chromosome-positive acute lymphoblastic leukemia. Ann Hematol. 2021; 100: 2479-86.
10.Yu CH, Lin TK, Jou ST, Lin CY, Lin KH, Lu MY, et al. MLPA and DNA index improve the molecular diagnosis of childhood B-cell acute lymphoblastic leukemia. Sci Rep. 2020; 10: 11501.
11.Mitchell RJ, Kirkwood AA, Barretta E, Clifton-Hadley L, Lawrie E, Lee S, et al. IKZF1 alterations are not associated with outcome in 498 adults with B-precursor ALL enrolled in the UKALL14 trial. Blood Adv. 2021; 5: 3322-32.
12.Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2006; 103: 6688-93.
13.Moorman AV, Barretta E, Butler ER, Ward EJ, Twentyman K, Kirkwood AA, et al. Prognostic impact of chromosomal abnormalities and copy number alterations in adult B-cell precursor acute lymphoblastic leukaemia: a UKALL14 study. Leukemia. 2022; 36: 625-36.
14.Hrabovsky S, Vrzalova Z, Stika J, Jelinkova H, Jarosova M, Navrkalova V, et al. Genomic landscape of B-other acute lymphoblastic leukemia in an adult retrospective cohort with a focus on BCR-ABL1-like subtype. Acta Oncol. 2021; 60: 760-70.
15.Yoon JH, Yhim HY, Kwak JY, Ahn JS, Yang DH, Lee JJ, et al. Minimal residual disease-based effect and long-term outcome of first-line dasatinib combined with chemotherapy for adult Philadelphia chromosome-positive acute lymphoblastic leukemia. Ann Oncol. 2016; 27: 1081-8.
Search
News